3D Sementic Segmentation on Electron Microscopy Dataset
Introduction
This passage will cover a experiment of sementic segmentation. The dataset includes a section of the Electron Microscopy Dataset fron EPFL represents a 5x5x5µm section taken from the CA1 hippocampus region of the brain, corresponding to a 1065x2048x1536 volume.
Setup
Dataset
The train and test dataset are both sub-volumns from the complete dataset. Each of them consists of 165 slices of the 1065x2048x1536 volume.
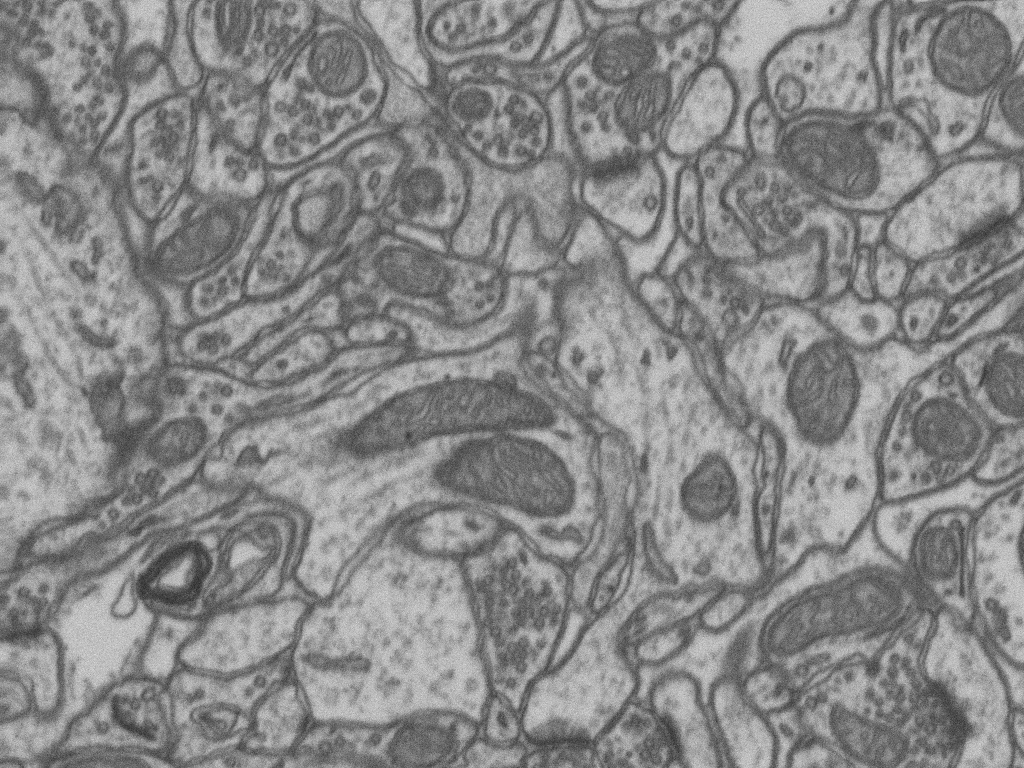
The groundtruth will annotate mitochondria in two sub-volumns.

Segmentation model
The model we use here is SegFormer: The SegFormer model was proposed in SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers by Enze Xie, Wenhai Wang, Zhiding Yu, Anima Anandkumar, Jose M. Alvarez, Ping Luo. The model consists of a hierarchical Transformer encoder and a lightweight all-MLP decode head to achieve great results on image segmentation benchmarks such as ADE20K and Cityscapes.
Data processing
- Download the
.tiffile of train and test splits from EPFL, open it, and create a dataset:
def read_tif(path):
images = []
with Image.open(path) as img:
for i in range(img.n_frames):
img.seek(i)
images.append(img.copy())
images = np.array(images)
return images
def create_dataset(image_path, label_path):
images = read_tif(image_path)
labels = read_tif(label_path)
dataset = Dataset.from_dict({"image": images,
"annotation": labels})
return dataset
train_path, train_label_path = os.path.join("data","training.tif"), os.path.join("data","training_groundtruth.tif")
test_path, test_label_path = os.path.join("data", "testing.tif"), os.path.join("data","testing_groundtruth.tif")
train_dataset = create_dataset(train_path, train_label_path)
test_dataset = create_dataset(test_path, test_label_path)
id2label = {0: 'background', 255: 'target'}
label2id = {'background':0, 'target': 255}
num_labels = len(id2label)
- Set transformation
checkpoint = "nvidia/mit-b0"
image_processor = AutoImageProcessor.from_pretrained(checkpoint, reduce_labels=False)
jitter = ColorJitter(brightness=0.25, contrast=0.25, saturation=0.25, hue=0.1)
def transforms(example_batch):
images = [jitter(x.convert("RGB")) for x in example_batch["image"]]
labels = [x for x in example_batch["annotation"]]
inputs = image_processor(images, labels)
return inputs
def val_transforms(example_batch):
images = [jitter(x.convert("RGB")) for x in example_batch["image"]]
labels = [x for x in example_batch["annotation"]]
inputs = image_processor(images, labels)
return inputs
train_dataset.set_transform(transforms)
test_dataset.set_transform(val_transforms)
- Implement evaluation metrices
def compute_metrics(eval_pred): with torch.no_grad(): logits, labels = eval_pred logits_tensor = torch.from_numpy(logits) logits_tensor = nn.functional.interpolate( logits_tensor, size=labels.shape[-2:], mode="bilinear", align_corners=False, ).argmax(dim=1) pred_labels = logits_tensor.detach().cpu().numpy() metrics = metric.compute( predictions=pred_labels, references=labels, num_labels=num_labels, ignore_index=255, reduce_labels=False, ) for key, value in metrics.items(): if isinstance(value, np.ndarray): metrics[key] = value.tolist() return metrics - load pretrained model
model = AutoModelForSemanticSegmentation.from_pretrained(checkpoint, id2label=id2label, label2id=label2id)
- Set training arguments, trainer, and start training
training_args = TrainingArguments( output_dir=os.path.join("data", "Electron-Microscopy-seg"), learning_rate=6e-5, num_train_epochs=50, per_device_train_batch_size=2, per_device_eval_batch_size=2, save_total_limit=3, evaluation_strategy="steps", save_strategy="steps", save_steps=20, eval_steps=20, logging_steps=10, eval_accumulation_steps=5, remove_unused_columns=False, ) trainer = Trainer( model=model, args=training_args, train_dataset=train_dataset, eval_dataset=test_dataset, compute_metrics=compute_metrics, ) trainer.train()
This is the basic training process, more details will be updated in the future :)